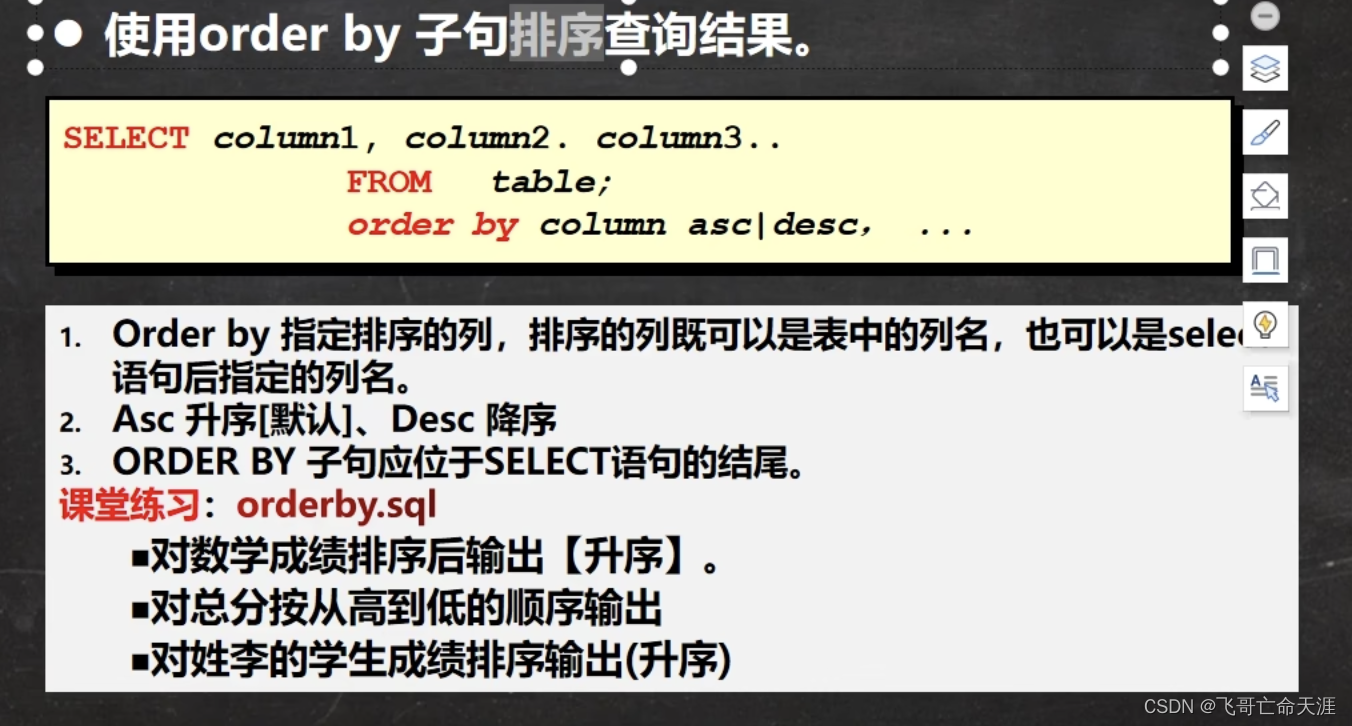
引用“using Clustering”,使用方法如下:
1. K-means
简单例子:
using Clustering
# make a random dataset with 1000 random 5-dimensional points
X = rand(5, 1000)
# cluster X into 20 clusters using K-means
R = kmeans(X, 20; maxiter=200, display=:iter)
@assert nclusters(R) == 20 # verify the number of clusters
a = assignments(R) # get the assignments of points to clusters
c = counts(R) # get the cluster sizes
M = R.centers # get the cluster centers
可选参数包括:
- init (defaults to :kmpp): how cluster seeds should be initialized, could be one of the following:
a Symbol, the name of a seeding algorithm (see Seeding for a list of supported methods); an instance of SeedingAlgorithm; an integer vector of length k that provides the indices of points to use as initial seeds. - weights: n-element vector of point weights (the cluster centers are the weighted means of cluster members)
- display::Symbol: the level of information to be displayed. It may take one of the following values:
:none: nothing is shown
:final: only shows a brief summary when the algorithm ends
:iter: shows the progress at each iteration
2. K-medoids
使用方式类似k-means,调用参数包括:
kmedoids!(dist::AbstractMatrix, medoids::Vector{Int};
[kwargs...]) -> KmedoidsResult
* medoids::Vector{Int}: the indices of k medoids
* assignments::Vector{Int}: the indices of clusters the points are assigned to, so that medoids[assignments[i]] is the index of the medoid for the i-th point
* costs::Vector{T}: assignment costs, i.e. costs[i] is the cost of assigning i-th point to its medoid
* counts::Vector{Int}: cluster sizes
* totalcost::Float64: total assignment cost (the sum of costs)
* iterations::Int: the number of executed algorithm iterations
* converged::Bool: whether the procedure converged
3. Hierarchical Clustering
使用方式类似k-means,调用参数包括:
hclust(d::AbstractMatrix; [linkage], [uplo], [branchorder]) -> Hclust
Arguments
* d::AbstractMatrix: the pairwise distance matrix.
* linkage::Symbol: cluster linkage function to use. linkage defines how the distances between the data points are aggregated into the distances between the clusters. Naturally, it affects what clusters are merged on each iteration. The valid choices are:
:single (the default): use the minimum distance between any of the cluster members
:average: use the mean distance between any of the cluster members
:complete: use the maximum distance between any of the members
:ward: the distance is the increase of the average squared distance of a point to its cluster centroid after merging the two clusters
:ward_presquared: same as :ward, but assumes that the distances in d are already squared.
* uplo::Symbol (optional): specifies whether the upper (:U) or the lower (:L) triangle of d should be used to get the distances. If not specified, the method expects d to be symmetric.
* branchorder::Symbol (optional): algorithm to order leaves and branches. The valid choices are:
:r (the default): ordering based on the node heights and the original elements order (compatible with R's hclust)
:barjoseph (or :optimal): branches are ordered to reduce the distance between neighboring leaves from separate branches using the "fast optimal leaf ordering" algorithm
Fields
* merges::Matrix{Int}: matrix encoding subtree merges:
each row specifies the left and right subtrees that are merged negative subtree id denotes the leaf node and corresponds to the data point at position −id positive id denotes nontrivial subtree (the row merges[id, :] specifies its left and right subtrees)
* linkage::Symbol: the name of cluster linkage function used to construct the hierarchy (see hclust)
* heights::Vector{T}: subtree heights, i.e. the distances between the left and right branches of each subtree calculated using the specified linkage
* order::Vector{Int}: the data point indices ordered so that there are no intersecting branches on the dendrogram plot. This ordering also puts the points of the same cluster close together.
4. DBSCAN
dbscan(points::AbstractMatrix, radius::Real,
[leafsize], [min_neighbors], [min_cluster_size]) -> Vector{DbscanCluster}
Cluster points using the DBSCAN (density-based spatial clustering of applications with noise) algorithm.
Arguments
points: matrix of points. points
radius::Real: query radius
Optional keyword arguments to control the algorithm:
leafsize::Int (defaults to 20): the number of points binned in each leaf node in the KDTree
min_neighbors::Int (defaults to 1): the minimum number of a core point neighbors
min_cluster_size::Int (defaults to 1): the minimum number of points in a valid cluster
Example
points = randn(3, 10000)
# DBSCAN clustering, clusters with less than 20 points will be discarded:
clusters = dbscan(points, 0.05, min_neighbors = 3, min_cluster_size = 20
5. Markov Cluster Algorithm
mcl(adj::AbstractMatrix; [kwargs...]) -> MCLResult
Perform MCL (Markov Cluster Algorithm) clustering using adjacency (points similarity) matrix.
Arguments
add_loops::Bool (enabled by default): whether the edges of weight 1.0 from the node to itself should be appended to the graph
expansion::Number (defaults to 2): MCL expansion constant
inflation::Number (defaults to 2): MCL inflation constant
save_final_matrix::Bool (disabled by default): whether to save the final equilibrium state in the mcl_adj field of the result; could provide useful diagnostic if the method doesn't converge
prune_tol::Number: pruning threshold
6. Affinity Propagation
affinityprop(S::AbstractMatrix; [maxiter=200], [tol=1e-6], [damp=0.5],
[display=:none]) -> AffinityPropResult
Perform affinity propagation clustering based on a similarity matrix S.
Sij is the similarity (or the negated distance) points, Sii defines the availability of the i-th point as an exemplar.
Arguments
damp::Real: the dampening coefficient, 0≤damp<1. Larger values indicate slower (and probably more stable) update. damp=0 disables dampening.
Fields
* exemplars::Vector{Int}: indices of exemplars (cluster centers)
* assignments::Vector{Int}: cluster assignments for each data point
* iterations::Int: number of iterations executed
* converged::Bool: converged or not
7. 模糊c均值
fuzzy_cmeans(data::AbstractMatrix, C::Int, fuzziness::Real,
[...]) -> FuzzyCMeansResult
Perform Fuzzy C-means clustering over the given data.
Arguments
data::AbstractMatrix: Each column represents one d-dimensional data point.
C::Int: the number of fuzzy clusters, 2≤C<n
fuzziness::Real: clusters fuzziness, fuzziness>1
Optional keyword arguments:
dist_metric::Metric (defaults to Euclidean): the Metric object that defines the distance between the data points
Fields
centers::Matrix{T}: matrix with columns being the centers of resulting fuzzy clusters
weights::Matrix{Float64}: matrix of assignment weights
iterations::Int: the number of executed algorithm iterations
converged::Bool: whether the procedure converged
Examples
using Clustering
# make a random dataset with 1000 points
# each point is a 5-dimensional vector
X = rand(5, 1000)
# performs Fuzzy C-means over X, trying to group them into 3 clusters
# with a fuzziness factor of 2. Set maximum number of iterations to 200
# set display to :iter, so it shows progressive info at each iteration
R = fuzzy_cmeans(X, 3, 2, maxiter=200, display=:iter)
# get the centers (i.e. weighted mean vectors)
# M is a 5x3 matrix
# M[:, k] is the center of the k-th cluster
M = R.centers
# get the point memberships over all the clusters
# memberships is a 20x3 matrix
memberships = R.weights